Is MS1 utilized in DIA-MS?
During a DIA-MS experiment, MS1 scans are usually acquired. For the TOF-based DIA-MS experiments, MS1 scans are usually of moderate resolution. For example, the original SWATH-MS paper (PMID: 22261725) by Gilet et al used 250-ms to finish a DDA-MS1 scan while 100-ms to finish a SWATH-MS1 on a TOF-MS system. Considering that each scan, regardless of MS1 or MS2 in SWATH, takes 0.1 s, SWATH-MS1 takes around 3.3% of the cycle time. The authors did, however, additionally described the SWATH-MS1 scan to be “optional” (figure below).
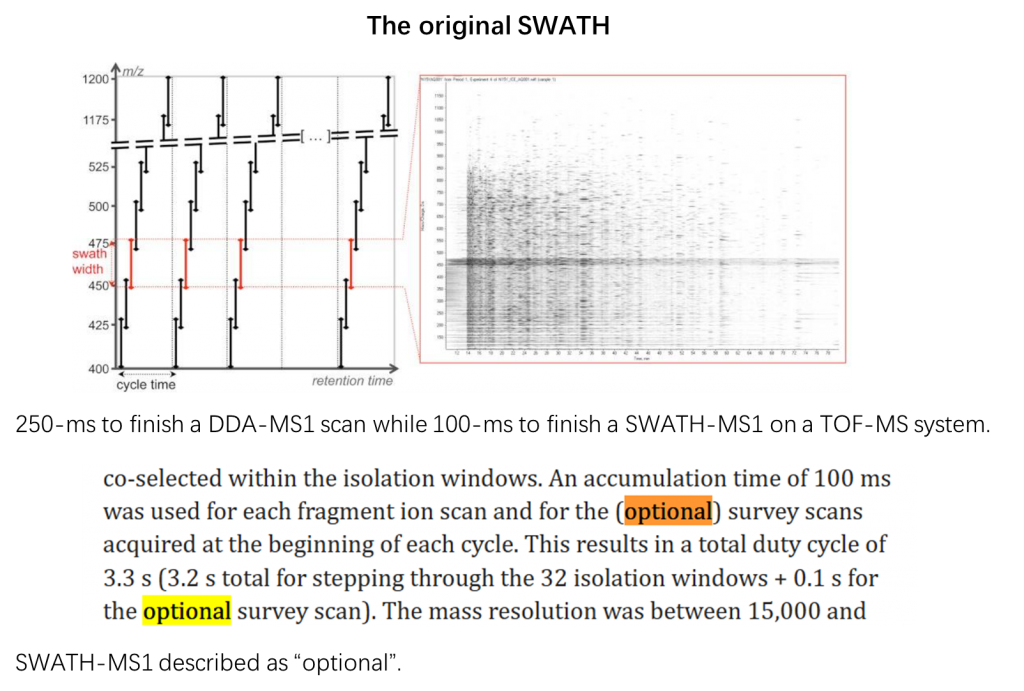
In the recent Scanning SWATH paper (PMID: 33767396), the design of a conventiomal SWATH was modified that the accumulation times of MS1 were shortened to 10 ms, even lower than the 25 ms for the SWATH MS/MS scans. Especially, MS1 scans were omitted when designing high-sensitivity Scanning SWATH. diaPASEF (PMID: 33257825) by Meier et al keeps MS1 but repeats two rounds of MS/MS acquisition in one cycle to limit the overall number of MS1 scans. Therefore, just as Rardin et al. commented in their MCP paper (PMID: 22261725), these TOF-based MS1 scans “are underutilized or even deemphasized by some vendors during DIA workflows”.
We noticed exceptions when DIA-MS was conducted on an Orbitrap system. AIF (all-ion fragmentation) (PMID: 20610777) developed in 2010 is the original Orbitrap-based DIA method that fragments precursors in the scan range as a whole without segmentation (figure below). MS1 and MS2 scans are set at the same resolution (100k) and accumulation time (1s), in another word, MS1 takes 50% of the cycle time. The evolved hyper-reaction monitoring (HRM) (PMID: 25724911) is a SWATH-type DIA method on Orbitrap and takes about 5% of the cycle time to collect a MS1 scan with the same resolution as MS2 (35k).

We came to the initial conclusion that 1 MS1 + n MS2 is a recognized DIA-MS paradigm. Empirically,orbitrap-based DIA-MS1 seems to be designed with higher qualities than TOF-based DIA-MS1.
Is MS1 utilized in DIA analysis?
There are two possible ways for a MS1 to be utilized in DIA data analysis.
Score chromatograph
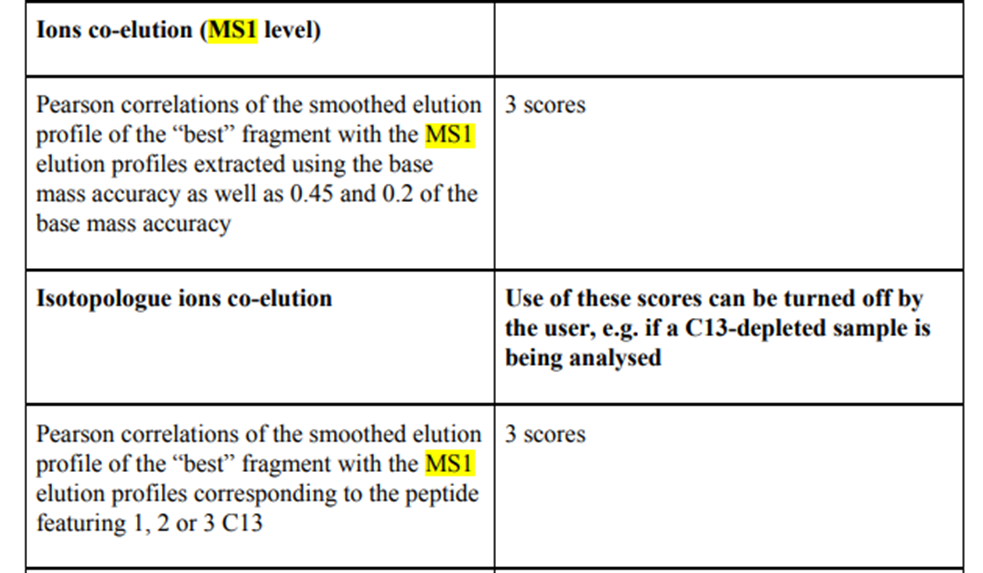
OpenSWATH is a primary software tool for SWATH/DIA analysis (PMID: 24727770), following an automated and targeted analysis fashion of the queried peptides. OpenSWATH has an option -use-ms1-trace that upon use will do a peak picking on the MS1 chromatographs and thereafter calculate a few (rudimentary) scores based on MS1 data. Same logic applies to DIA-NN (PMID: 31768060), wherein seven MS1-related scores are used (figure below).
MS1-level quantification
MS1-level quantification involves extracting ion currents (usually three isotopologues) from MS1, correcting interferences (optional), and calculating the covered area for intensities. Current software tools that enable MS1-level quantification include Spectronaut, DIA-Umpire, Integrated Dual Scan Analysis in Skyline, and MaxDIA. MS1-based quantification in Skyline is discouraged in Skyline.
Empirically, MS1 is comprehensively used for identification but less popularly used in quantification.
How to improve DIA analysis with MS1?
Bruderer et al (PMID: 29070702) found that increasing the resolution of MS1 from 30k to 120k while maintaining the cycle time, although necessarily decreases the MS2 window number, increases total peptide identification. This is attributed to the enhanced confidence of peptides based on their MS1-level scores. They also found that increasing MS1 resolution is helpful for MS1-based quantification, although it does not outperform MS2-based quantification.
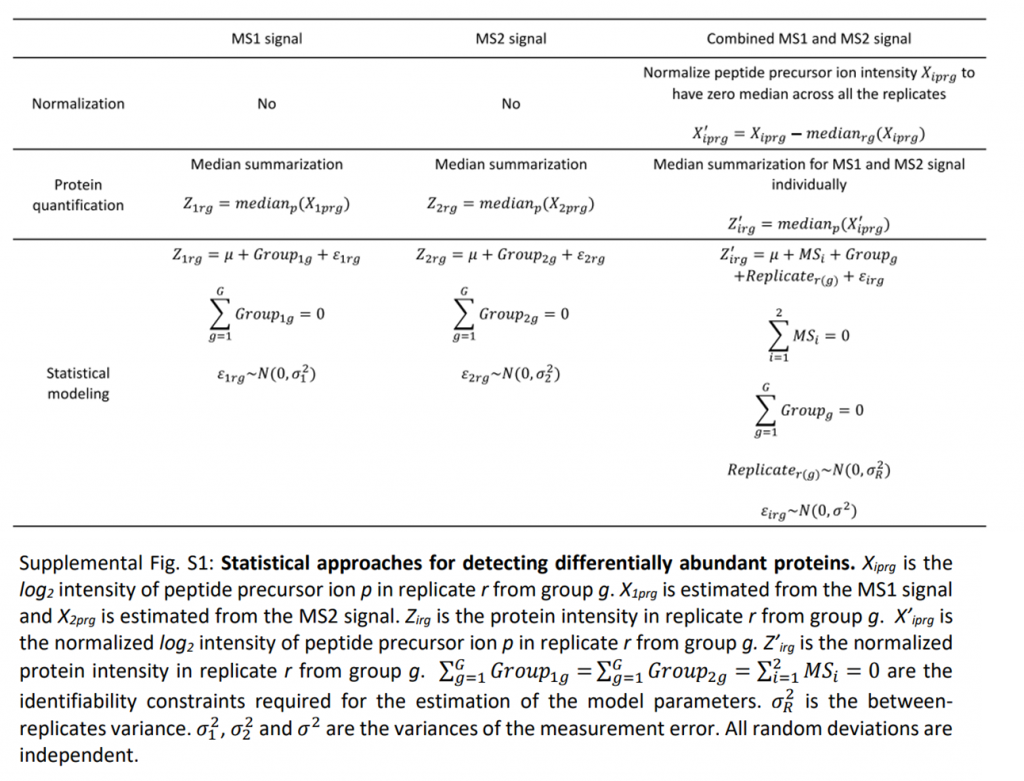
Huang et al (PMID: 31888964) combined MS1-level and MS2-level quantification results via a statistical approach shown below. This hybrid method could increase confidences of differential analysis, both identificationally and quantitationally, especially for the less confident proteins with marginally significant fold changes. maxDIA is also capable of hybrid quantification, although details are not elaborated in the main text.
We came to the conclusion that a high-quality MS1 increases ID. A hybrid (MS1+MS2) quantification increases quantification.
Discussion
We now know that taking MS1 into consideration in DIA-MS analysis is necessary to enhance peptide identification, and helps increase quantification performance when combined with MS1-level results. DIA methods that emphasize MS1, such as Boxcarmax (PMID: 33533601), will be of great promise if enlengthened total run time is not an issue of concern.
However, we do not know if sacrificing MS1 to allow for more cycles, when the total run time are limited, will bring to better results, especially quantificationally, since more data points can be collected to generate a nicer peptide chromatograph.