Note: The analyses were originated from citexs.com and I made several adjustments on the layout and content. For academic use only.
1.摘要
本文基于PubMed文献数据库,使用citexs赛特新思数据分析平台,对2012-01至2022-12关于 single cell 和 proteomics 研究进行文献分析及大数据挖掘,梳理该领域的发展历程,分析研究主题的热点变化等,将分析结果可视化,旨在对single cell 和 proteomics相关领域研究进行系统综述和趋势展望,探讨当前的研究动态及热点,挖掘分析尚需改进的内容,以期为后续研究提供参考。
2.数据来源
(1)数据来源于PubMed数据库
(2)时间跨度:2012-01至2022-12
(3)文献类型:article
(4)语种:English
(5)共检索到 1443 篇文献
(6)检索词:(single cell) AND (proteomics)
3.研究方法
本文采用文献计量学方法,以PubMed文献数据库为基础,使用citexs赛特新思数据分析平台,进行文献大数据挖掘,分别选择国家、机构、作者、关键词、期刊、关联疾病、关联基因等对象进行数据挖掘和分析,可视化呈现该领域的总趋势、分布情况、热点变化等。
4.1 年度发文趋势分析
以single cell 和 proteomics为关键词搜索,2012-01至2022-12的文献有 1443 篇,文献年均发文量132篇。如图1所示,2021达到年发文量顶峰310篇,2020增长率最快为42.33%,提示该领域的研究得到快速发展,处于快速上升阶段。
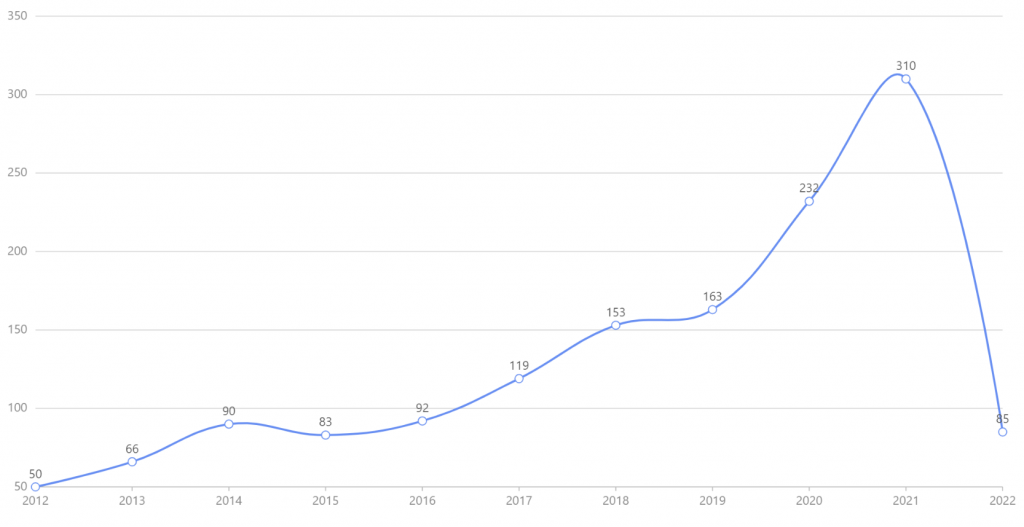
图1. 2012-01至2022-12 single cell 和 proteomics相关文献的年度发文趋势
4.2 研究国家分析
2012-01至2022-12,全球在 single cell 和 proteomics研究领域发文量前64的国家如图2所示, 在该领域发文量最多的国家是United States of America(606篇,42%),China(181篇,12.54%)和Germany(180篇,12.47%)位居第二和第三。
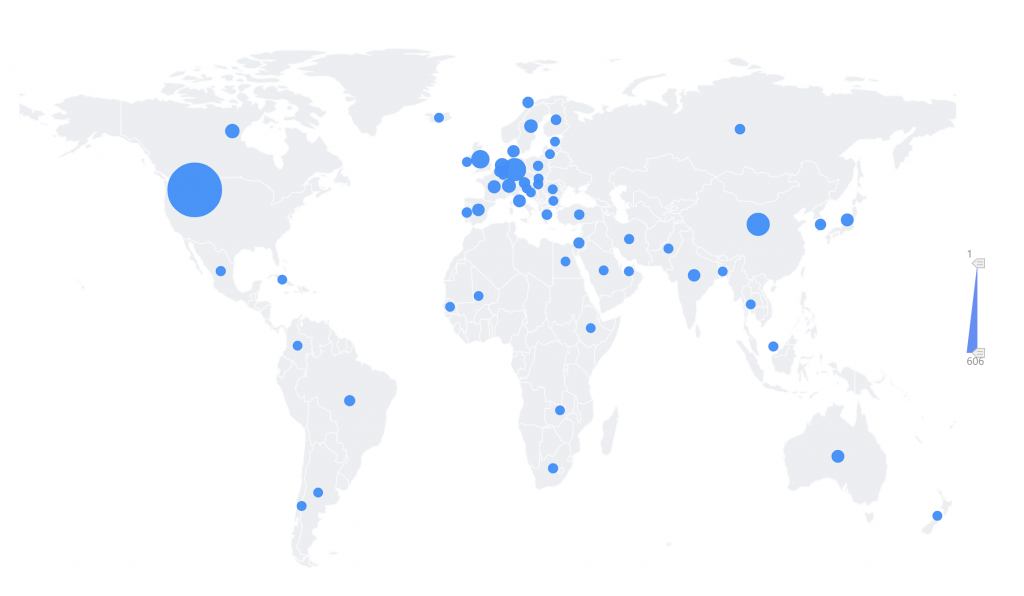
图2. 2012-01至2022-12 single cell 和 proteomics的研究国家分析
4.3 研究机构分析
2012-01至2022-12,全球在 single cell 和 proteomics 研究领域发文量前20的国家研究机构如图3所示,其中university of california和university of cambridge发文量占据前两名,分别发表了48篇和21篇,university of copenhagen发表了21篇,位于第三名。
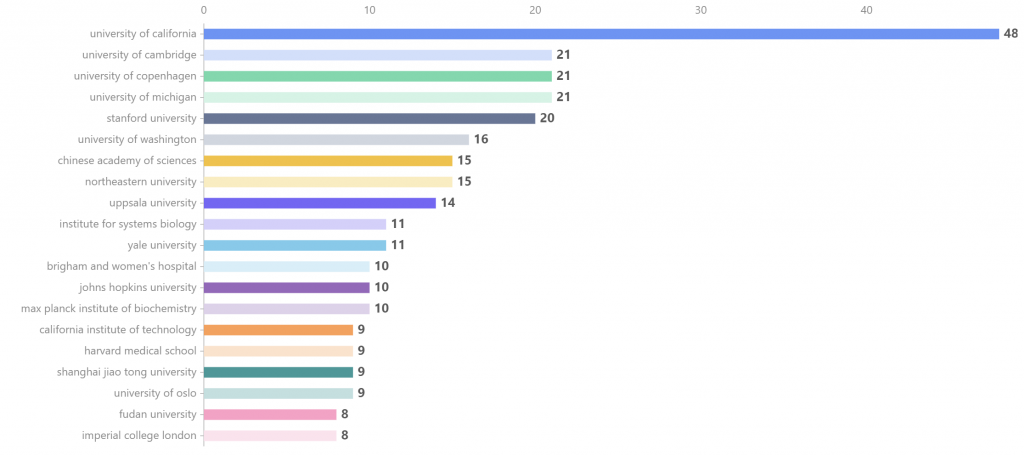
图3. 2012-01至2022-12 single cell 和 proteomics的研究机构分析
4.4 研究作者分析
2012-01至2022-12,全球在 single cell 和 proteomics 研究领域发文量前30的作者见表1。该领域产出文献最多的作者是Richard D Smith,迄今为止一共发表21篇文献;Ying Zhu位居第二,发表了19篇文献;Ronald J Moore位居第三,已有14篇文献发表。 (声明:文献原始数据来源于pubmed,由于其部分机构缺失和部分杂志未收录,造成文章数量偏少,排名仅供参考!)
| Rank | Author | Institute&country | No. |
| 1 | Richard D Smith | united states | 21 |
| 2 | Ying Zhu | united states | 19 |
| 3 | Ronald J Moore | united states | 14 |
| 4 | Wei-jun Qian | united states | 13 |
| 5 | Rui Zhao | united states | 12 |
| 6 | Tao Liu | united states | 12 |
| 7 | Nikolai Slavov | northeastern university, united states | 11 |
| 8 | Cecilia Lindskog | uppsala university, sweden | 10 |
| 9 | William B Chrisler | united states | 9 |
| 10 | Chia-feng Tsai | united states | 8 |
| 11 | Jing Zhou | united states | 7 |
| 12 | Ryan T Kelly | united states | 7 |
| 13 | Tujin Shi | united states | 7 |
| 14 | Angus I Lamond | university of dundee, united kingdom | 6 |
| 15 | Anil K Shukla | united states | 5 |
| 16 | Masanori Aikawa | brigham and women’s hospital, united states | 5 |
| 17 | Ryan T Kelly | brigham young university, united states | 5 |
| 18 | Fabian J Theis | institute of computational biology, germany | 4 |
| 19 | Jesper V Olsen | university of copenhagen, denmark | 4 |
| 20 | Ling Zhong | university of new south wales, australia | 4 |
| 21 | Mark J Raftery | university of new south wales, australia | 4 |
| 22 | Matthias Mann | max planck institute of biochemistry, germany | 4 |
| 23 | Paul D Piehowski | united states | 4 |
| 24 | Romain Huguet | united states | 4 |
| 25 | Rong Fan | yale university, united states | 4 |
| 26 | Sean Mackay | united states | 4 |
| 27 | Alexander Schmidt | university of basel, switzerland | 3 |
| 28 | Daniel Lopez-ferrer | united states | 3 |
| 29 | Fredrik Pontén | uppsala university, sweden | 3 |
| 30 | Joshua N Adkins | united states | 3 |
4.5 发文期刊分析
2012-01至2022-12,以single cell 和 proteomics为关键词搜索,检索出论文1443篇,发文量排名前30的期刊见图4,其中刊载文献量最多的期刊是J Proteome Res(84篇);Mol Cell Proteomics位居第二,刊载文献量60篇;Methods Mol Biol位居第三,刊载文献量54篇。
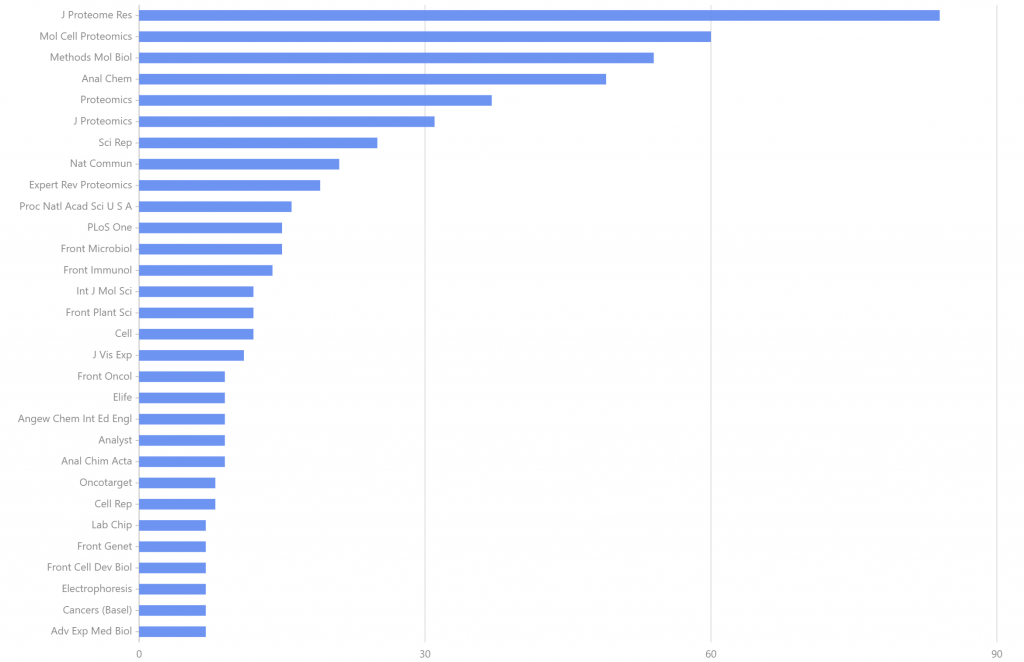
图4. 2012-01至2022-12 single cell 和 proteomics的发文期刊分析
4.6 关键词热点词频分析
论文关键词是对研究目的、研究对象、研究方法进行高度凝练与概括。基于关键词的分析能够反映某一研究领域某一时间段内主题演变趋势和研究热点。把single cell 和 proteomics作为为关键词搜索,时间跨度为2012-01至2022-12,如图5所示,出现频次前5的关键词分别是: proteomics,mass spectrometry,transcriptomics,metabolomics,single-cell proteomics.
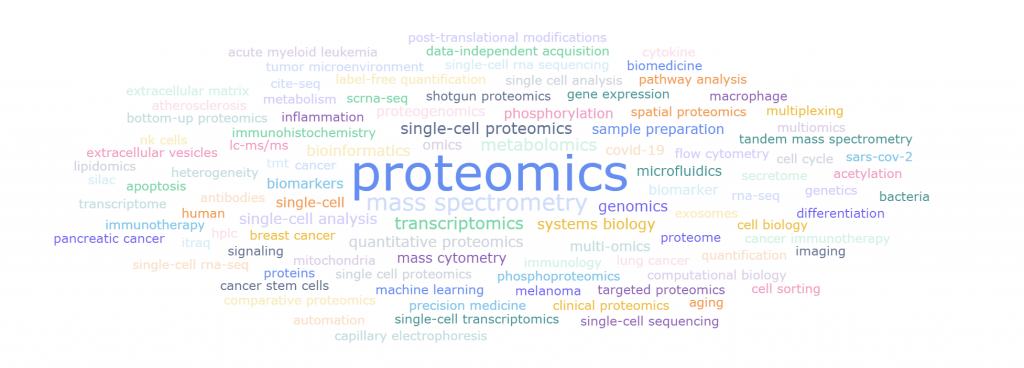
图5. 2012-01至2022-12 single cell 和 proteomics的热点词频分析
4.7 关键词热度随时间变化分析
将2012-01至2022-12分为4个时间段,如图6表示与single cell 和 proteomics相关联的关键词词频在各个时间段的热度排名及排名变化情况。
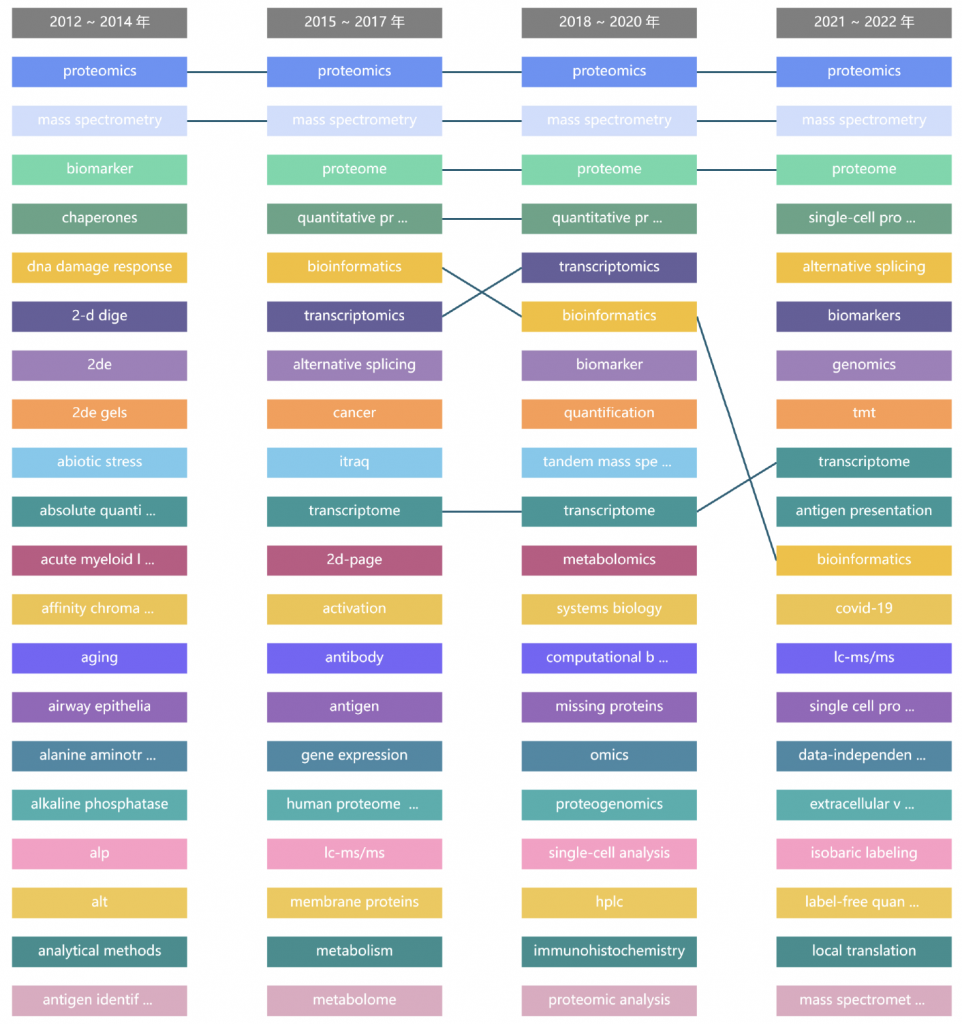
图6. 2012-01至2022-12 single cell 和 proteomics不同时间段的热度排名及排名变化分析
4.8 关联基因分析
2012-01至2022-12,以single cell 和 proteomics为关键词搜索,检索出论文1443篇,如图7所示,其中文献量最多的TP53(95篇);EGFR位居第二,文献量为71篇;AKT1位居第三,文献量66篇。
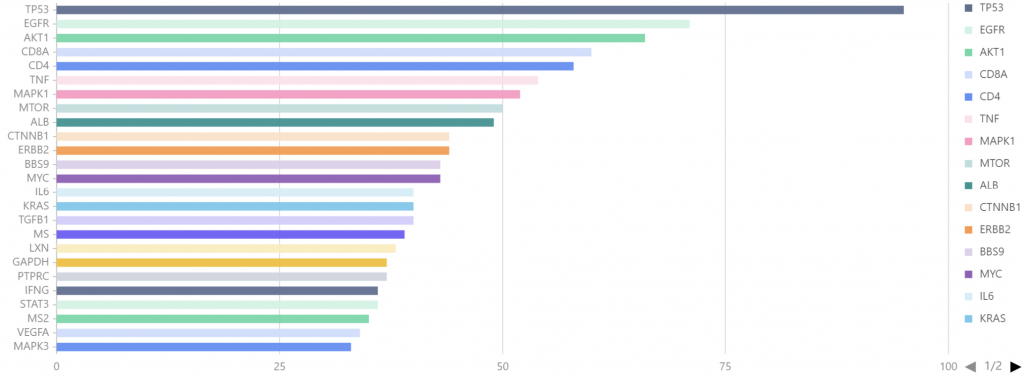
图7. 2012-01至2022-12 single cell 和 proteomics关联基因分析
5.1 AI推荐文献
根据搜索关键词single cell 和 proteomics,综合文献被引用数、发表时间、年份、影响因子等条件,推荐出30篇文献,见表2。
| Title | Journal | IF | PMID | |
| 1 | Single-cell proteomics | Nat Methods | 28.547 | 30573843 |
| 2 | A dream of single-cell proteomics | Nat Methods | 28.547 | 31406385 |
| 3 | Defining the carrier proteome limit for single-cell proteomics | Nat Methods | 28.547 | 33288958 |
| 4 | Single-Cell Proteomics | Trends Biochem Sci | 33653632 | |
| 5 | In Situ Single Cell Proteomics Reveals Circulating Tumor Cell Heterogeneity during Treatment | ACS Nano | 15.881 | 34225455 |
| 6 | Scaling Up Single-Cell Proteomics | Mol Cell Proteomics | 34808355 | |
| 7 | Multiplexed single-cell proteomics using SCoPE2 | Nat Protoc | 34716448 | |
| 8 | Single-cell proteomics takes centre stage | Nature | 34545225 | |
| 9 | Ultrasensitive single-cell proteomics workflow identifies >1000 protein groups per mammalian cell | Chem Sci | 9.825 | 34163866 |
| 10 | Single-Cell Proteomics Reveal that Quantitative Changes in Co-expressed Lineage-Specific Transcription Factors Determine Cell Fate | Cell Stem Cell | 24.633 | 30880026 |
| 11 | Single-Cell Proteomics for Cancer Immunotherapy | Adv Cancer Res | 6.242 | 29941105 |
| 12 | Single-cell Proteomics: Progress and Prospects | Mol Cell Proteomics | 32847821 | |
| 13 | Driving Single Cell Proteomics Forward with Innovation | J Proteome Res | 34597050 | |
| 14 | Single-cell proteomics reveals changes in expression during hair-cell development | Elife | 8.140 | 31682227 |
| 15 | Transformative Opportunities for Single-Cell Proteomics | J Proteome Res | 4.466 | 29945450 |
| 16 | Single-cell proteomics: A treasure trove in neurobiology | Biochim Biophys Acta Proteins Proteom | 33845200 | |
| 17 | Mass Spectrometry-Based Analytical Strategy for Single-Cell Proteomics | Methods Mol Biol | 34905166 | |
| 18 | Features of Peptide Fragmentation Spectra in Single-Cell Proteomics | J Proteome Res | 34920664 | |
| 19 | Quantitative Accuracy and Precision in Multiplexed Single-Cell Proteomics | Anal Chem | 34967612 | |
| 20 | Spatial proteomics: a powerful discovery tool for cell biology | Nat Rev Mol Cell Biol | 94.444 | 30659282 |
| 21 | Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type-specific immune regulation in atopic dermatitis | J Allergy Clin Immunol | 10.793 | 32344053 |
| 22 | Microfluidic Single-Cell Proteomics Assay Chip: Lung Cancer Cell Line Case Study | Micromachines (Basel) | 34683198 | |
| 23 | Advancing single-cell proteomics and metabolomics with microfluidic technologies | Analyst | 4.616 | 30351310 |
| 24 | Replication of single-cell proteomics data reveals important computational challenges | Expert Rev Proteomics | 34602016 | |
| 25 | Single Cell Proteomics Using Multiplexed Isobaric Labeling for Mass Spectrometric Analysis | Methods Mol Biol | 34766268 | |
| 26 | In-Depth Mass Spectrometry-Based Single-Cell and Nanoscale Proteomics | Methods Mol Biol | 33165848 | |
| 27 | A deeper look at carrier proteome effects for single-cell proteomics | Commun Biol | 35194133 | |
| 28 | Imaging the future: the emerging era of single-cell spatial proteomics | FEBS J | 33351222 | |
| 29 | An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics | Nat Commun | 14.919 | 30814501 |
| 30 | Microchip-based single-cell functional proteomics for biomedical applications | Lab Chip | 6.799 | 28280819 |
5.2 经典文献
基于AI推荐文献共引用关系筛选出经典文献30篇,见表3。
| Title | Journal | IF | PMID | |
| 1 | Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents | Mol Cell Proteomics | 5.911 | 15385600 |
| 2 | Cytoscape: a software environment for integrated models of biomolecular interaction networks | Genome Res | 9.043 | 14597658 |
| 3 | Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources | Nat Protoc | 19131956 | |
| 4 | Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips | Nat Protoc | 13.491 | 17703201 |
| 5 | Large-scale analysis of the yeast proteome by multidimensional protein identification technology | Nat Biotechnol | 54.908 | 11231557 |
| 6 | Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS | Anal Chem | 6.986 | 12713048 |
| 7 | Global quantification of mammalian gene expression control | Nature | 21593866 | |
| 8 | Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells | Proc Natl Acad Sci U S A | 22203961 | |
| 9 | The Perseus computational platform for comprehensive analysis of (prote)omics data | Nat Methods | 28.547 | 27348712 |
| 10 | MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification | Nat Biotechnol | 54.908 | 19029910 |
| 11 | Mass spectrometry-based proteomics | Nature | 49.962 | 12634793 |
| 12 | Nanodroplet processing platform for deep and quantitative proteome profiling of 10-100 mammalian cells | Nat Commun | 14.919 | 29491378 |
| 13 | Ultrasensitive proteome analysis using paramagnetic bead technology | Mol Syst Biol | 11.429 | 25358341 |
| 14 | Proteomics. Tissue-based map of the human proteome | Science | 47.728 | 25613900 |
| 15 | Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets | Cell | 41.582 | 26000488 |
| 16 | Andromeda: a peptide search engine integrated into the MaxQuant environment | J Proteome Res | 4.466 | 21254760 |
| 17 | Skyline: an open source document editor for creating and analyzing targeted proteomics experiments | Bioinformatics | 20147306 | |
| 18 | Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry | Nat Methods | 24584193 | |
| 19 | Universal sample preparation method for proteome analysis | Nat Methods | 19377485 | |
| 20 | Single-Cell Mass Spectrometry for Discovery Proteomics: Quantifying Translational Cell Heterogeneity in the 16-Cell Frog (Xenopus) Embryo | Angew Chem Int Ed Engl | 15.336 | 26756663 |
| 21 | Mass Cytometry: Single Cells, Many Features | Cell | 27153492 | |
| 22 | SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation | Genome Biol | 13.583 | 30343672 |
| 23 | Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum | Science | 21551058 | |
| 24 | Quantitative analysis of complex protein mixtures using isotope-coded affinity tags | Nat Biotechnol | 54.908 | 10504701 |
| 25 | Single-cell western blotting | Nat Methods | 24880876 | |
| 26 | The PRIDE database and related tools and resources in 2019: improving support for quantification data | Nucleic Acids Res | 16.971 | 30395289 |
| 27 | The MaxQuant computational platform for mass spectrometry-based shotgun proteomics | Nat Protoc | 13.491 | 27809316 |
| 28 | Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics | Mol Cell Proteomics | 5.911 | 12118079 |
| 29 | An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database | J Am Soc Mass Spectrom | 24226387 | |
| 30 | Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ | Mol Cell Proteomics | 24942700 |